| Michael W. Hunkapiller |
 |
|
 |
 |
 |
|

[figure 7]

[figure 8]
[figure 9]

[figure 10]

[figure 11]

[figure 12]

[figure 13]

[figure 14]

[figure 15] |
The initial
advance that we made in Lee Hood's lab was the realization that
if you went away from one color, or one band, on a gel and instead
moved to a detection system that used a different color for each
of the four reactions, then you could combine all the reactions
together, process them on a single strip of gel, and more clearly
determine the order that they were coming through on the gel. It
was this fluorescence labeling concept and, in particular, the ability
to use four different fluorescent labels, each paired with one of
the nucleic acid component types, that really gave rise to most
of the modern day forms of the Sanger sequencing methodology. [figure
7] Additional methods, shown here, use labels attached to what is
called a "primer," the small bit of DNA molecule that is used to
start the synthesis and copying of these fragments to generate the
different series of sizes of DNA that are analyzed.
The first implementation of that came out in 1986 in the form of
the "Model 370 DNA Sequencer." [figure 8] The model 370 was built
as an automated sequencer, but to be honest, it was more semi-automated
than automated. The automated part involved the use of a spectroscopic
technique, as opposed to visually reading an autoradiogram picture,
for directly inputting the data into a computer. The computer then
interpreted the sequence using fairly strict algorithms, which decreased
the degree of ambiguity previously associated with the process.
The downside was that sequencing was still a slow process. It relied
on slab-gels; it relied on the art of people making those gels;
and it relied upon people manually loading samples into the sequencer.
Nevertheless, it did allow for the successful study of a lot of
DNA.
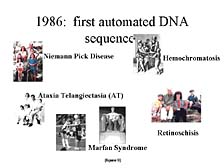
Although the throughput of DNA sequencers present in 1986 was limited,
they did enable researchers to sequence and look at fairly small
bits of DNA, e.g., a single gene or a collection of a few genes.
[figure 9] As limited as this was, it allowed for the elucidation
of the association between very specific genes and inherited diseases
that had pretty dark consequences for the people who were unfortunate
enough to inherit them. Researchers were able to show the slight
variations in the DNA sequence of key genes that caused some of
these problems.
As the technology improved over the next several years, the throughput
of the systems began to increase substantially. [figure 10] The
first thing that one could do was begin to look at the genomes (whole
set of genes)of very small organisms. Viruses were more easily tackled
initially than other organisms, but eventually, "free-living" organisms,
those containing enough genome components to carry out all the biological
processes they need to function, began to happen in the mid-1990's.
In particular, Craig and his team at The Institute for Genomic Research
(TIGR) successfully sequenced the first free-living organism, Haemophilus
influenzae, in the mid-1990's. I am sure Craig will talk about this
a little more later.
We realized that if you began to fully automate the process -- if
you took away a lot of the work associated with preparing gels and
loading samples and allowed the systems to be run around the clock
-- that it gave rise to the possibility of sequencing an organism
as complicated as the human being (whose genome contains 3 billion
base pairs) in a relatively short period of time. While we had developed
the first capillary-based instrument in 1995, it was designed to
prove the principles of doing capillary electrophoresis DNA sequencing,
as opposed to the traditional slab-gel technique pioneered by Sanger's
group. [figure 11] However, it was the model 3700 DNA sequencer
and the collection of technologies that went into it -- some that
we developed, some as Dr. Matsubara pointed out, that were developed
in collaboration with Hideki Kambara at Hitachi and Norm Dovichi
at the University of Alberta -- that allowed us to take the same
kind of full automation that we had developed in a very low-throughput
single-capillary system and put it into a high-throughput multi-capillary
unit. The 3700 had the kind of sensitivity and high-throughput that
allowed genome-wide studies in a very short amount of time.
The culmination of this technology development, in one sense, was
the dual publication of the first drafts of the human genome in
Science, by Craig's group at Celera, and in Nature, by the publicly
funded Human Genome Project, in February of this past year. [figure
12] This was the first time these drafts were presented to the scientific
world. The instrument that I showed on the previous slide was used
to generate about 90 percent of the total amount of sequence data
that went into the assemblies of the human genome in the 12 months
prior to publication.
When you ask what enabled this to happen in such a short period
of time, it's important to realize that this wasn't just an engineering
problem of applying the technology that had evolved over the 15
years since our first automated system. [figure 13] Rather, it was
the entire process of determining how to strategically select which
DNA samples to sequence, how to generate these samples from a complex
organism, how to prepare that DNA for analysis, how to know what
kind of enzymes to use to carry out the Sanger sequencing reactions,
which methodology to employ to attach fluorescent labels, as well
as the mechanics of separating the DNA in a high-throughput fashion
and then interpreting these fragments using a very complex informatics
system, and putting the fragments together again. I will not discuss
a lot of these, but what I would like to emphasize is that being
recognized for this work, as Craig is, we both have extremely large
teams below us that did most of the work. [figure 14] This is a
list of the people who were on the 3700 development team. They come
from a diverse set of disciplines -- chemistry, biochemistry, molecular
biology, mechanical and electrical engineering, software and firmware
development, as well as the manufacturing of both the chemicals
used in the process and the instrumentation itself. Thus, the development
of technology like this and the provision of this technology to
the world, as a research tool for use on a routine basis, is a complex
endeavor that really requires an enormous variety of skills to carry
it out successfully.
One of the key innovations in going from the initial, partially
automated systems to the 3700 was that, for the first time, robotics
were employed in loading samples into the system for analysis, thereby
cutting down on the labor requirements and the number of errors
associated with mis-applying samples that confuse very large projects.
[figure 15] This concept of automation, which cut the labor requirements
by more than 90 percent, allowed the concept of large factory output
(by using a large number of these machines) to be applied to complex
projects like sequencing the human genome. |
|
|