| Stephen P.A. Fodor |
 |
|
 |
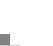 |
 |
|

[Slide 11]

[Slide 12]
[Slide 13]

[Slide 14]

[Slide 15]

[Slide 16]

[Slide 17]
[Slide 18]
[Slide 19]
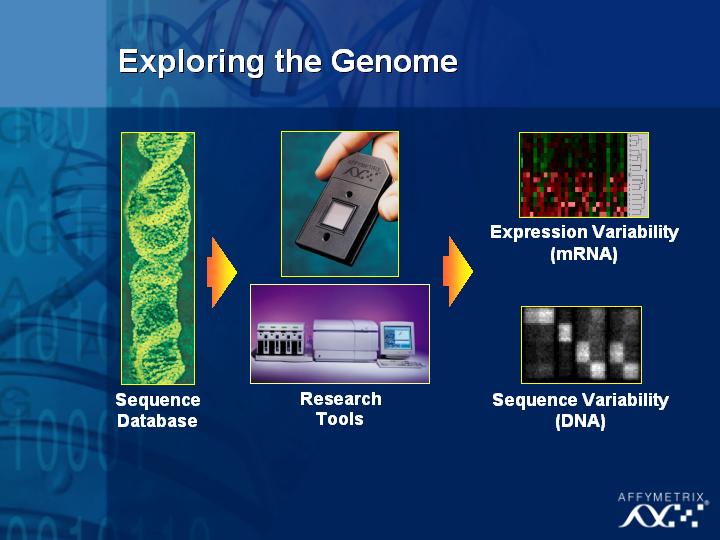
[Slide 20]
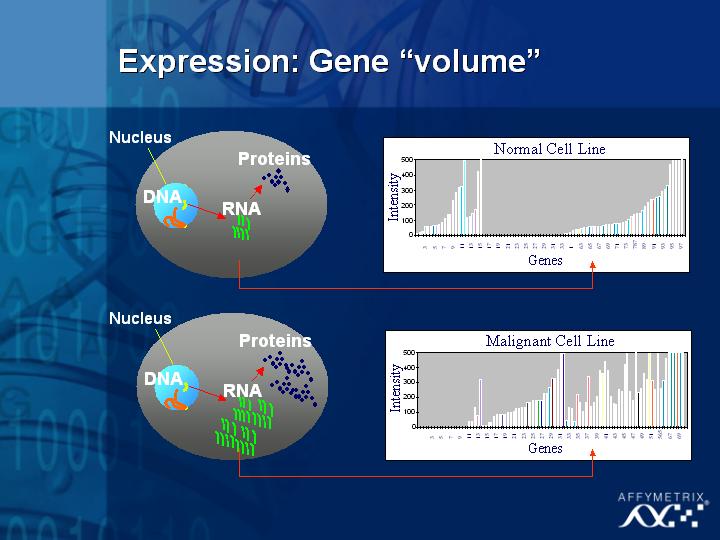
[Slide 21]

[Slide 22]
|
[Slide 11]
We use light as a chemical reagent to activate the wafer's surface for additional chemical reactions.
[Slide 12,13]
The wafers are then cut into individual chips, packaged and shipped to customers.
[Slide 14,15]
These arrays can be manufactured in different formats corresponding to the amount of genetic content required.
[Slide 16]
Affymetrix' current commercial microarrays contain approximately 500,000 features each containing a different DNA sequence.
[Slide 17]
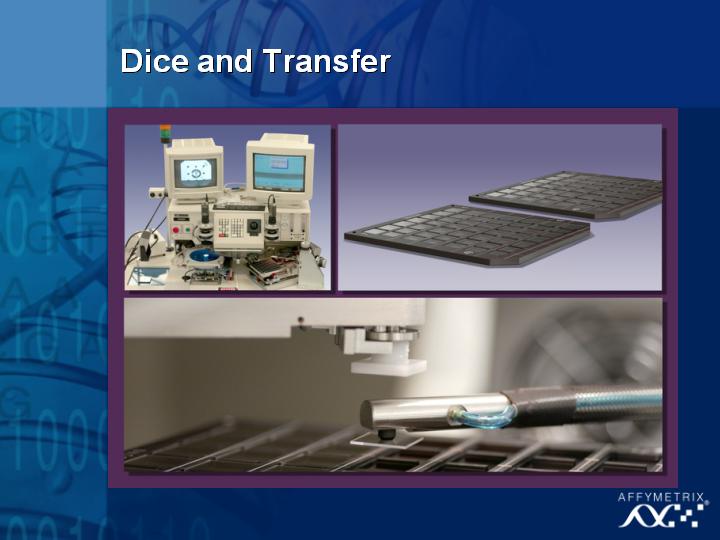
In 1989, we only had one system, located at our headquarters in Silicon Valley, California, the birthplace of microarray technology.
[Slide 18]
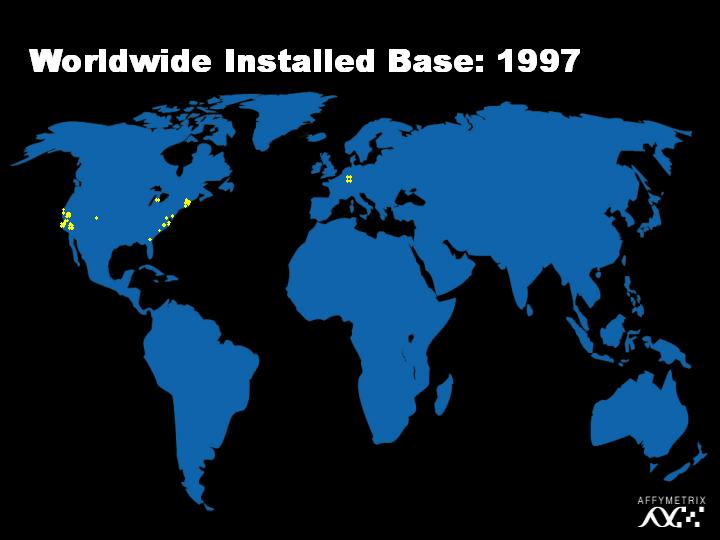
However, the field grew quickly and in 1997, we saw a number of academic research centers and pharmaceutical companies adopting the technology.
[Slide 19]
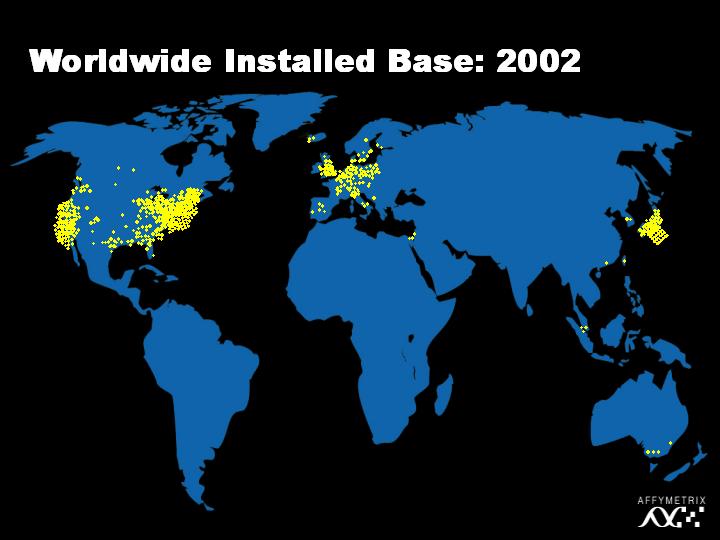
And now in 2002, there are literally thousands of investigators using this technology. To put this into perspective, in the last three months, we shipped around one hundred thousand chips with five hundred thousand features on them. That is roughly fifty billion DNA data points sold in a period of three months.
[Slide 20]
The two major areas that this technology has been applied to are the understanding of expression variations and of DNA sequence variations.
[Slide 21]
I will spend some time talking about each of these subjects. The first is gene expression, understanding which genes are turned on or off in different samples.
[Slide 22]
Today, the most popular microarray set we produce is called the Human GeneChip U133 set, which consists of two chips, which harbor about one million DNA sequences and interrogate around 33,000 different transcripts of the human genome. We have taken the commercial perspective of making all the probe set sequences available, as well as mapping them to the draft of the human genome. Of course this type of approach can be used for many other organisms, but today I will focus primarily on humans.
|
|
|