| Stephen P.A. Fodor |
 |
|
 |
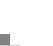 |
 |
|
[Slide 23]
[Slide 24]
[Slide 25]
[Slide 26]
[Slide 27]
[Slide 28]

[Slide 29]
[Slide 30]
[Slide 31]
[Slide 32]
[Slide 33]

[Slide 34]
[Slide 35]
|
[Slide 23]
In the pharmaceutical and biotechnology industries, these products are used heavily in all different aspects of the drug discovery and development process. Historically, this technology was used primarily in the early stages of research and development, but more recently these products have moved downstream into pre-clinical, Phase I, Phase II, and Phase III safety studies. The goal of this research is of course to make the process of moving new compounds from research into downstream clinical testing much more efficient and less risky.
[Slide 24]
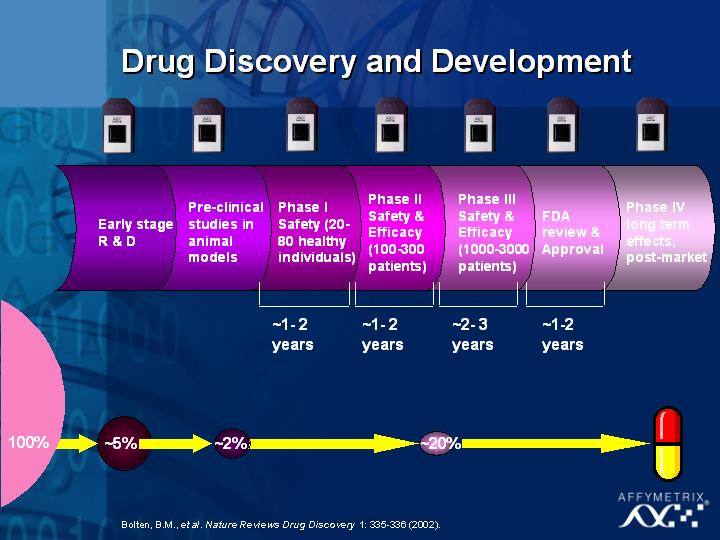
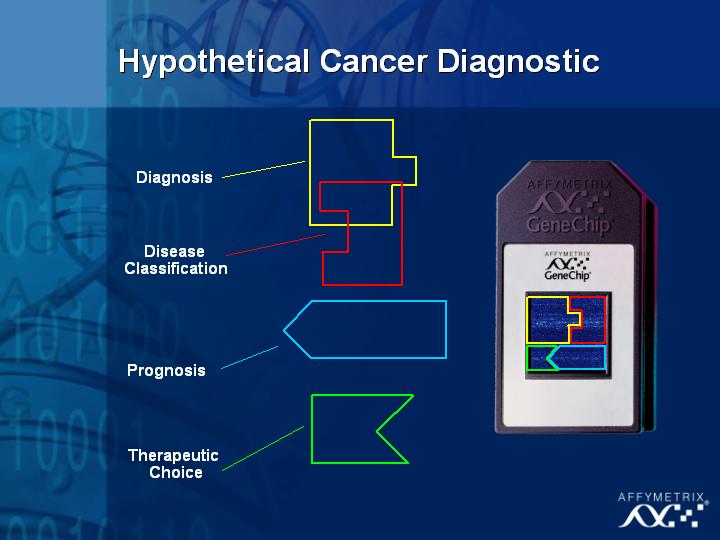
Another research area that has received a lot of attention, and I am sure we will hear more about this in the next talk, is the use of microarrays in understanding the molecular signatures of cancer. This is a direct application of the technology that will have a major impact on issues involving personalized medicine and treatment. Using microarrays, we can ask very specific questions about diagnosis, classification, prognosis and therapeutic choice in complex diseases such as cancer.
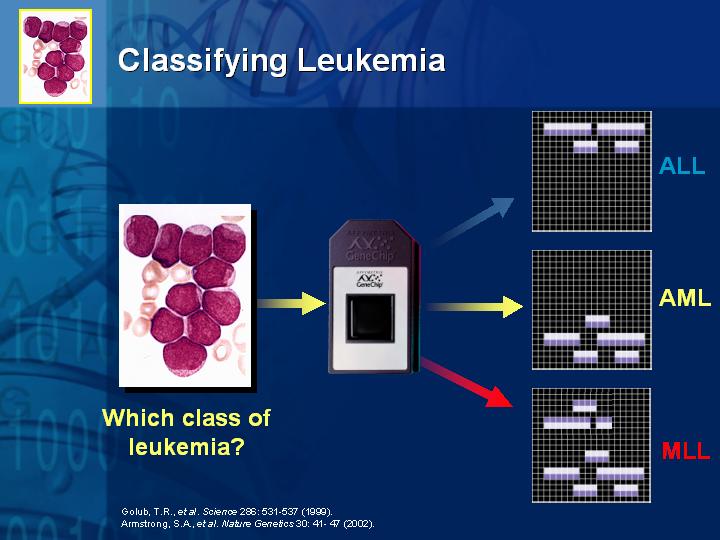
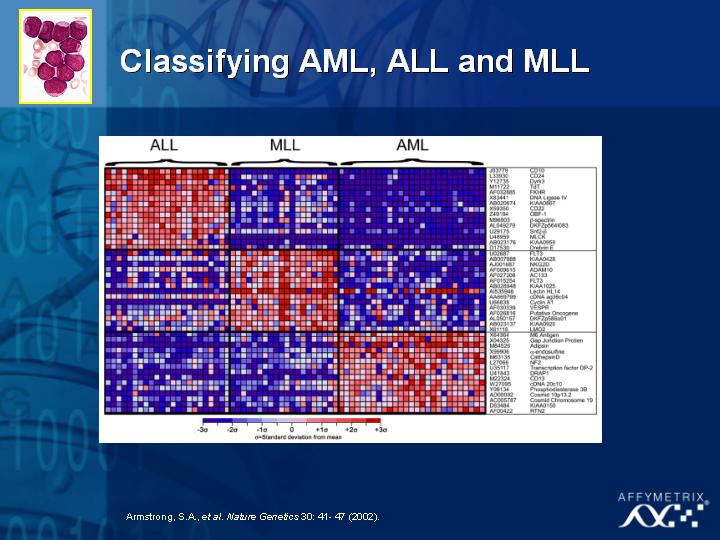
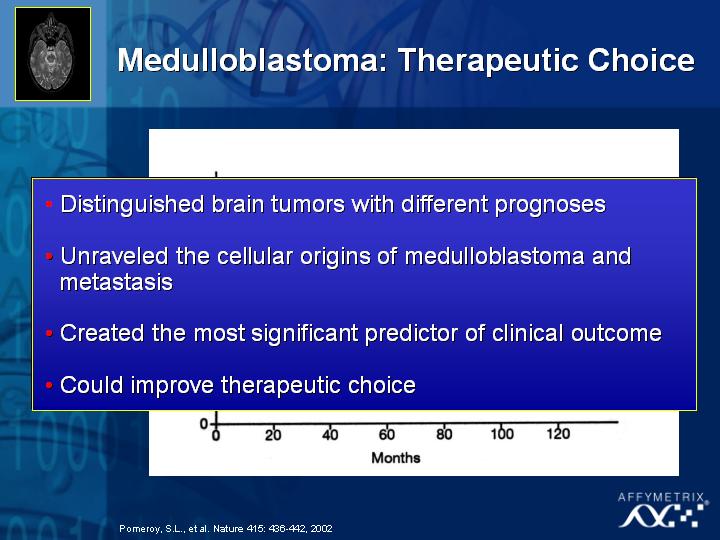
[Slide 25,26,27,28]
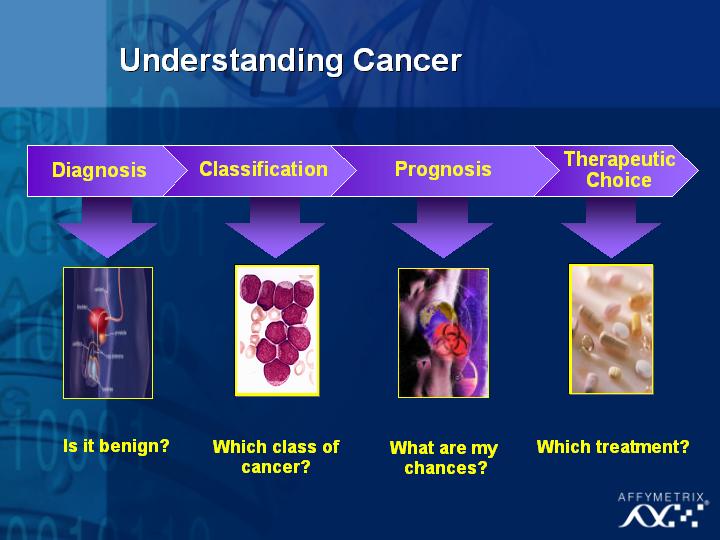
To date, the technology has already been used in diagnosing prostate cancer (Slide 25), in classifying new and different types of leukemia (Slide 26,27) and in childhood medulloblastoma (Slide 28) where some treatment decisions need to be made based on quality of life issues for the patients. The objective of this type of research is to distinguish the tumors with different prognoses, and ultimately improve the therapeutic choice for these patients.
[Slide 29]
There are a number of different scientific publications that have come out of in the literature on this topic, including those from my colleague, Dr. Brown.
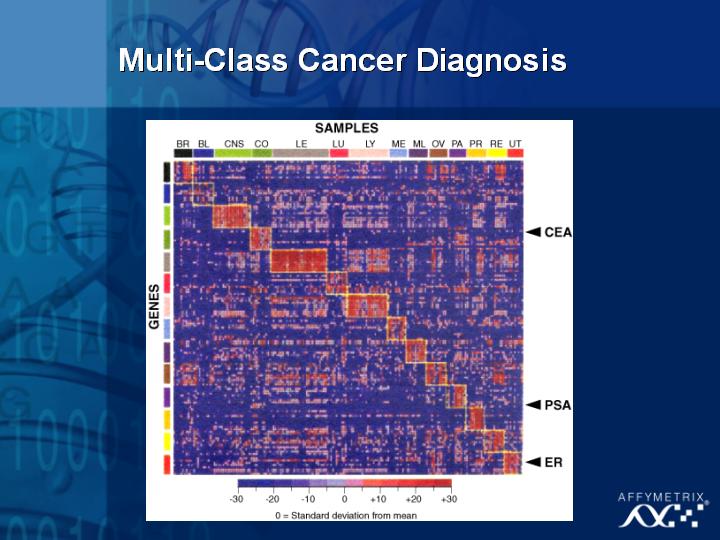
[Slide 30]
There are now studies to suggest that we may be able to create chips to look at many different genes and sub-classify different types of cancers.
[Slide 31]
This raises the possibility of creating one chip that can provide both, diagnostic and prognostic information.
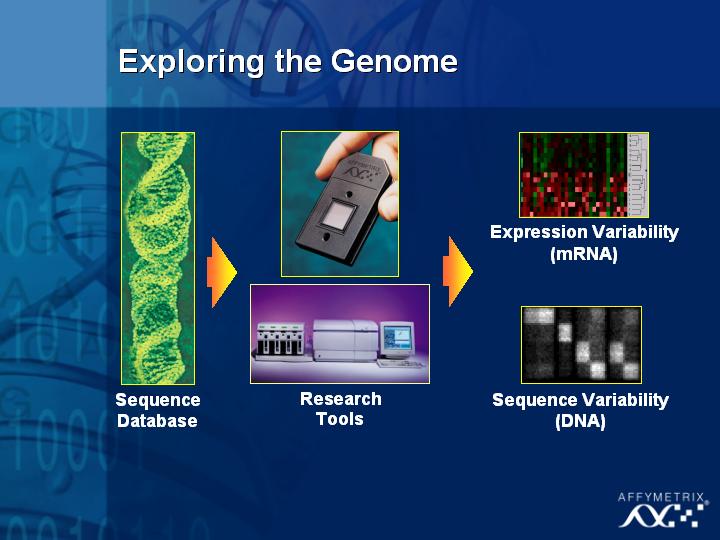
[Slide 32]
The second area I would like to touch briefly upon is the issue of DNA sequence variation.
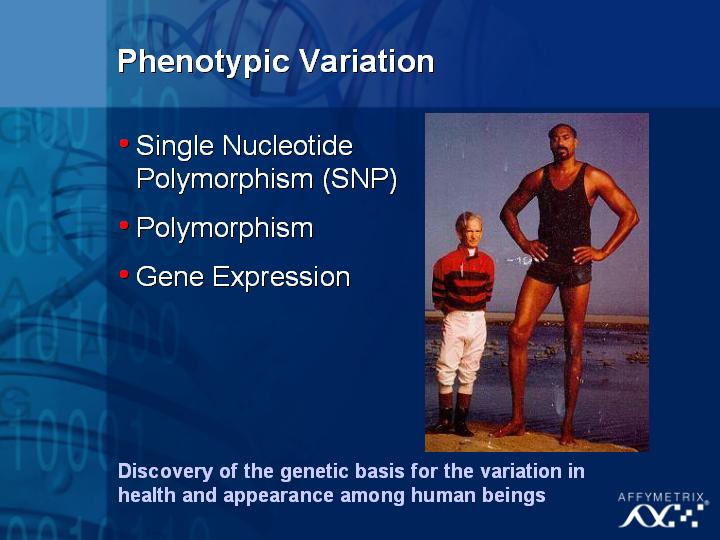
[Slide 33]
This is a very important topic, since we are all composed of basically the same genetic material. Although we share the same genome, if we can really understand the genetic basis of variation, we can understand many of the genetic differences between us. Ultimately, what we would like to do is to relate the variations of the human genome sequence to its effects on gene expression.
[Slide 34]
Furthermore, if we are able to understand genetic variation, we hope to some day stratify the population into groups that show a good response, no response, or perhaps even have a toxic response to particular therapeutic, with an ultimate goal of getting the right drug to the right person at the right time.
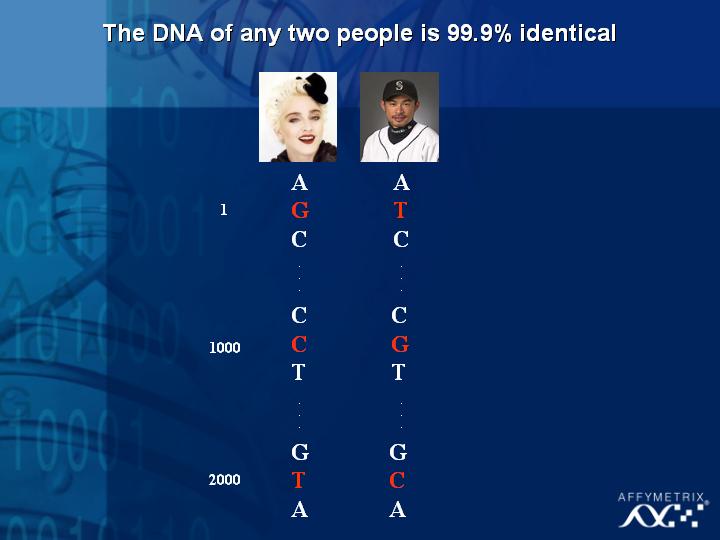
[Slide 35]
You might ask the question, what does Madonna and Ichiro have in common? Of course, the DNA in both of individuals is 99.9 % same. However, I think we all acknowledge that there are some significant differences between them. In fact, if we can begin to understand the very slight differences at varying positions in the genome perhaps we can begin to understand the differences between the broad spectrum of the population.
|
|
|