| Patrick O. Brown |
 |
|
 |
 |
 |
|

[Slide 1]
[Slide 2]

[Slide 3]
|
[Slide 1]
Thank you very much, Dr. Matsubara, and thanks to the people of the Takeda Foundation, Dr. Takeda, and the committee that selected me to share this great honor with Dr. Fodor.
It's a great honor and privilege to be here today, and I must say what a really wonderful treat it was to hear Dr. Fodor's terrific talk about his work.
I am going to begin my talk this morning, just as Steve did, by telling you first a little bit about the microarray technology that we developed, and then something about how my laboratory and other laboratories around the world are using DNA microarray technology in their research.
Then as an example I am going to tell you a little bit about a study that my laboratory has recently been doing, of one particular medical problem, stomach cancer, using DNA microarrays.
And finally, I'll share some thoughts about the future of this technology, and say something about the way that my group made this technology available for people to be able to choose.
When I began working on this project, it was based on an idea that I had for new way to study the problem that Steve talked about very eloquently, which was how differences our genes can produce the amazing variation in the human population that you can see just by looking around this room.
To make this idea work, it was critical to have a very easy and very inexpensive way to look at every human gene at once.
Like Dr. Fodor, I realized that the solution would be a tiny array of many thousands of DNA sequences - a DNA microarray.
[Slide 2]
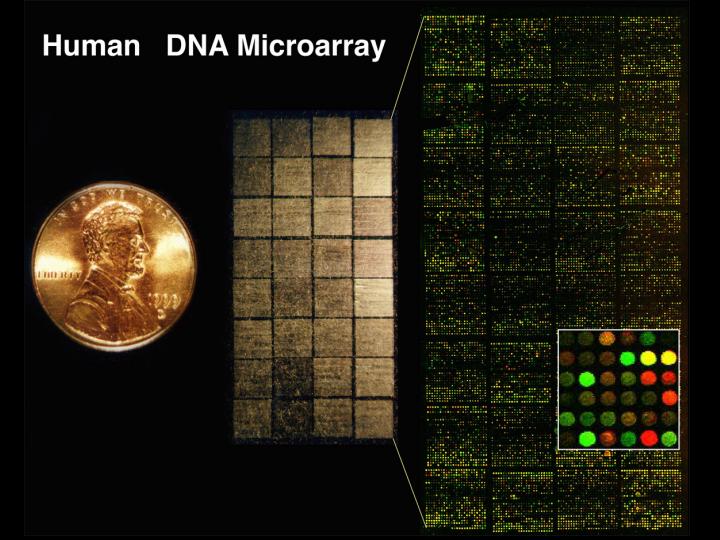
This is a picture of a DNA microarray that we use to look at human genome.
In this slide, this coin you can see on the left, is about the same size as a one Yen Japanese coin. And on left here you can see a microarray just as looks in visible light, before we use it in an experiment, all the little tiny sparkles you can see on this slide, around 40,000 of them, represent 33,000 different human genes. On the right, you can see the way that the array looks as one uses it in an experiment.
I think every scientist wants their work not only to be useful, but if possible, also to be beautiful and fun.
So, in designing this technology from the very beginning, I wanted a microarray experiment to be beautiful to look at, and I had a picture in my mind that the results would appear as a pattern of red and yellow and green dots. The patterns would change depending on the properties of the genes we would study.
Because our analysis uses a mixture of two samples, of human DNA or RNA - I won't go into the details of how we do it, but one of the samples is labeled with a green fluorescent dye and one labeled with a red fluorescent dye - the spots on the array change from these colorless sparkles to a pattern of red, yellow and green dots when we do a hybridization to analyze the samples.
The color of each spot represents a quantitative measurement of some property of the corresponding gene. In the example that I am showing in this slide, the color of each spot represents how active that particular gene is in a kind of cancer cell, a lymphoma. A red color, this spot, shows us that this gene is very active in this kind of cancer, and a green color, this spot, shows that this gene is turned off in this cancer.
So, we learn a lot about this cancer from the patterns of colors we see in these experiments and we also get a pretty picture out of it.
Of course, we don't analyze the spots just by looking at them. We use a digital imaging system to turn a color in a quantitative measurement. That'll be described just in the moment.
To make a microarray, instead using light and chemistry, the way that Steve's wonderful invention does, we took a much simpler and actually very ancient approach, which was simply to print DNA sequences on glass slides, just like you would print ink.
[Slide 3]
So we built this very simple robot to print a tiny drops of DNA onto the glass slide on this platform here. This particular robot was built by this wonderful graduate student in my lab at that time, Joe Derisi.
I think all you have to do is to compare this slide to some of the slides that Steve showed, and you can see how much less sophisticated and clever this technology is than Steve's method, but the simplicity has some benefits. The robot was very inexpensive and very easy to build - two people could put one of these together from parts out of a box at a cost of about 4 million yen in about one day.
The simplicity and the low cost made this technology accessible to ordinary biology labs with regular grant budgets. Another advantage of this very simple design is its flexibility. We can use this robot to print essentially any kind of molecule in liquid solution, including not only DNAs but also proteins for proteomics studies and even small molecules for studying chemical and biological interactions.
By keeping our microarrays very simple, user-friendly and cheap, we can make sure that the students who are using them in their research would not be afraid to use them to do adventurous experiments, all kinds of exploratory experiments to look at new questions about biology; and even people who are not professional scientists could use them to explore interesting questions of biology.
|
|
|