| Patrick O. Brown |
 |
|
 |
 |
 |
|

[Slide 12]
[Slide 13]
[Slide 14]
[Slide 15]
|
[Slide 12]
The biochemistry, as I said, suggests some ideas for how this enzyme might be acting to suppress progression of gastric cancer. One idea is that the protective effect is related to the role this enzyme can play in releasing arachidonic acid from cell membranes. The arachidonic acid can activate a particular programmed cell death pathway in carcinoma cells. If that mechanism is involved in the protective action of PLA2 group2A, then there's an immediate and very attractive possibility for treatment. We already have very non-toxic and widely used drugs, aspirin and non-steroid anti-inflammatory agents, that can provide another way to increase levels of arachidonic acid by blocking the cyclooxygenases that metabolize this molecule into prostaglandin and other derivatives.
In this regard, it's intriguing that there is epidemiological evidence that patients who were taking aspirin and NSAIDs, for example for arthritis, have a significantly reduced incidence of colon cancer and gastric cancer, which supports this possible model for the action.
The second interesting possibility comes from the fact that this particular phospholipase A2, out of the 15 different PLA2 enzymes encoded by the human genome, is unique in having a very potent anti-bacterial activity. That raises a possibility that bacteria have a role in progression of gastric cancer, in addition to their known role in initiating development of gastric cancer.
So, both of these models are exciting because they suggest at least possibility that there might be some very non-toxic therapies for treating patients of gastric cancer, particularly those who have low levels of PLA2 activity. One would be orally ingestion of this very stable enzyme itself and another is aspirin or NSAIDs, and the third possibility, if the anti-bacterial model were correct, would be antibiotic treatment.
Obviously, these are just some ideas that we are going to need to investigate further.
[Slide 13]
The microarray technology has only been around, certainly only been in widespread use for less than 10 years, so its potential applications are just beginning to be explored. You heard from Steve a wonderful vision for a diversity of applications of this technology. I'll just mention a few additional ideas about the near future of this technolody.
First of all, I think we're only just beginning to explore the human genome's expression program and how it gives rise to the diversity of cell types and cell functions in our body. I think we'll see the explosion of information over the next several years from this kind of basic biological study.
Secondly, I think it's very likely that over the next few years we are going to start to see actual applications in the clinic, using DNA microarrays in gene expression profiling for diagnosis of cancer and other diseases, and already there's quite lot of work underway moving in that direction. Particularly with the development of molecularly targeted therapies, I think this is going to be an important component of individualized treatments.
One thing that I didn't have a chance to talk about today but I think it's going to be a very big growth area in the use of DNA microarrays is to study and to monitor changes in our environment, like oceans, rivers, and soils, due to human activities, natural phenomena, and climate change, by monitoring and profiling living organisms, especially the microbial populations that are native to those environments.
Finally, I think we'll see a lot more use of microarrays in proteomic studies to study variation and changes in proteins - inside cells and also in body fluid like blood and urine - including a lot of effort in developing clinical applications like blood tests for cancer and other diseases.
[Slide 14]
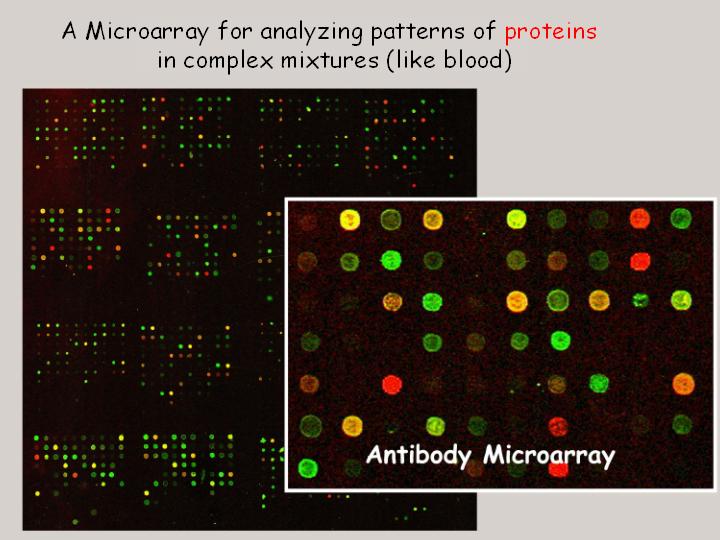
This slide shows an early version of an antibody microarray, a microarray in which the spots are composed of antibodies, which are used to measure variation in proteins in blood samples. I think we should be able to develop this technology to measure thousands of different proteins in a single drop of blood or in a small sample of cells. This work is still in progress, but I think it has lots of potential.
Dr. Fodor's work is a spectacular example of combining the invention of new technology with entrepreneurship in building a successful business to manufacture and commercialize the technology, in that way make it widely available for people to use.
[Slide 15]
I have no talent for business and not much interest in it. Instead, I work at the university where my job is basically to teach and carry out a basic research, which is about the best job in the world, actually.
I had always thought of our DNA microarray technology as a process rather than a product. So, for me the natural approach to distributing it wasn't to manufacture and sell it, which is something that I've not qualified to do anyway, but to teach others how to do it.
People in my lab, particularly Joe Derisi and Vishy Iyer, worked very hard with me and have been continuing to work on their own, to put together a complete set of instructions for building the simple robot that we use to print our microarrays as well as detailed protocols for using it to print the arrays and to carry out the experiments with them. Other members of my lab, particularly Mike Eisen, who's now running an independent lab of his own, developed and distributed free software for analysis of our microarray data.
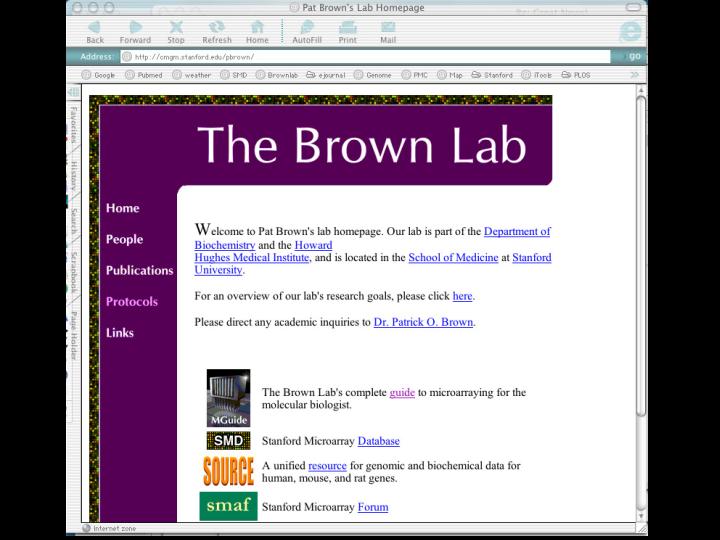
We made them available to people just by posting them on our lab website, where anyone could download them for free. This slide shows a view of our lab website and here is a link you can go to get the manual for how to make microarrays and the other sources for softwares and data and so forth like that.
This actually has worked very well and the cost is nothing to do this, since we are not manufacturing anything and we didn't have to ship anything, so we don't have to charge for it. I am sure, some days Steve wishes he had done this, so he wouldn't have to answer the questions about his pricing strategy.
The way we distributed our technology is just one example of the obvious and tremendous potential from the Internet to make a great volume of information available for free, at almost no cost to the producer, anywhere in the whole world. I think that this has tremendous and so far really undeveloped potential for bringing the benefits of scientific research in discovery to anyone who might be able to use them, scientists and physicians anywhere in the world, engineers, business people and also non-professionals who are just curious about science and technology and interested in learning more.
|
|
|