| Patrick O. Brown |
 |
|
 |
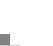 |
 |
|

[Slide 4]

[Slide 5]
[Slide 6]
[Slide 7]
|
[Slide 4]
So you can see the lady in this picture is about to put a sample on a microarray that she is holding in her hand there.
[Slide 5]
As you heard from Steve, microarrays can be used to study almost any property of a gene or genome. By far the most productive and widespread use, at least over the past few years, has been to study gene expression patterns, the scripts that our genome writes for each cell of our bodies.
[Slide 6]
There are thousands of different kinds of cells that make up our bodies, and they come in an incredible diversity of sizes and shapes, and have an extraordinary range of physiological properties and behaviors. This simple illustration just gives you an idea of some of this diversity. For example, you have cells like a neuron here, that has thousands of branches that make contact and exchange information with other cells. The branches can be up to a meter long. Then you have cells like the white blood cells that are tiny by comparison, move around in our blood, and are specialized to recognize and respond to infection or injury.
Every cell of your body has a complete copy of the genome, and the genome itself is a sort of a brain of that cell. It dictates how the cell is designed and how the cell can behave. All these cells that are so extraordinarily different from one another have identical genomes, completely identical set of genes. How can that be?
One way of looking at the answer is that the genes are like the words in the genome's vocabulary. All the cells use the same vocabulary, just as all the Japanese-speaking people here share the common vocabulary. And just as you can use the same vocabulary to write millions of different stories, the genome in each cell can use the same set of genes to write a different script for the life story for each different cell in your body.
One of the things that make these scripts so fascinating is that in every cell in your body right now the genome in that cell is writing a new page of a script, responding to the consequently changing conditions. So, it is a dynamic process, in which the gene expression pattern in each cell responds to changing conditions, and therefore studying those patterns can tell us about the conditions in which each cell finds itself.
Because the scripts are written in the language of genes using RNA transcripts of the genes, we can use the microarrays to read those scripts - to read the gene expression patterns of the cells and tissues of our bodies.
The biggest challenge we face in reading those scripts is that they are written in the genomes' own strange language, which is a language that we have very begun to learn. So, it is a challenge for us to try to understand the gene expression patterns that we see, when we study a new cell or tissue.
Our strategy is to try to learn the language by studying its patterns just the way a young child learns a language by immersing himself in it and observing very carefully all the ways that the each word is used.
[Slide 7]
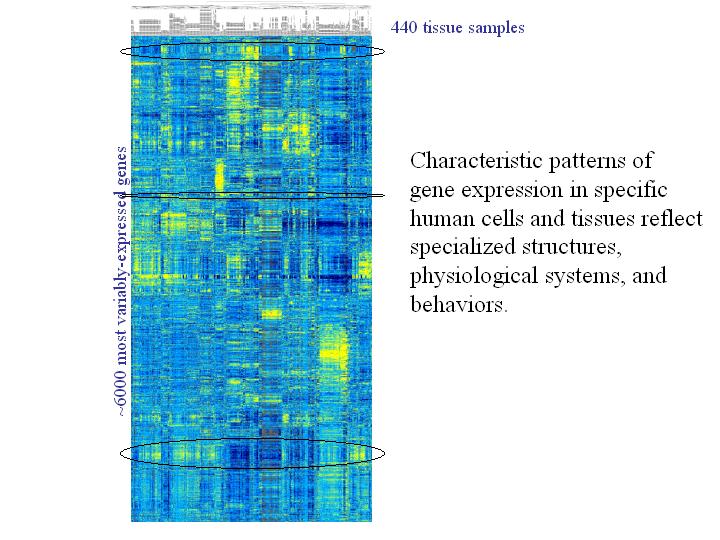
This slide illustrates one of the approaches we use for studying the patterns in the genome's language. You can think of it as a kind of a map of genome's expression patterns.
This map is organized just like a table, where each vertical column represents a set of measurements of expression of all the genes - in this case, 6,000 different genes - in a single sample, so each vertical column would be the results from the single microarray analysis.
And then each row represents a set of measurement of a single gene's expression in each of the different samples we analyzed. Each of, in this case, slightly more than 400 samples that were analyzed.
Each individual point or pixel on this map represents the measurement of one gene's expression in one particular tissue sample.
Nobody wants to try to make any sense of the data by reading the table with millions of numbers like this, and so it is much easier and more informative to represent the quantitative information using a color scale. And, in this case, we use a bright yellow color to represent the highest level of the expression, and a dark blue color to represent cases where genes did not express at all, and intermediate colors to represent intermediate levels of expression.
Then finally, to turn this into something like a useful map, we organized the data by clustering together the genes with most similar patterns of the expression and also we can also cluster together the cell or tissue samples that have the most similar expression patterns.
One thing that you see right away from looking at a picture like this is, on the one hand, how complex and intricate the genome's expression pattern is. How much variation there is among the genome's expression patterns and among cells in tissues in the particular set of genes that is expressed and their levels of expression. On the other hand, how highly orderly the program is. You see systematic groups of genes that are characteristically expressed together, and you can find cells with many different patterns of similarity and differences in their expression programs.
So we use these kinds of maps to see relationships between genes that tend to be characteristically expressed together and therefore likely to be involved in closely related functions, and to recognize the relationships between biological samples, different cells or tissue types that share common molecular features, and to relate specific genes to specific characteristics in the cell and tissue samples.
|
|
|